SVM Classifier for IRIS Data Set
Steps:
- Import the library files
- Read the dataset (Iris Dataset) and analyze the data
- Preprocessing the data
- Divide the data into Training and Testing
- Build the model - SVM Classifier with different types of kernels
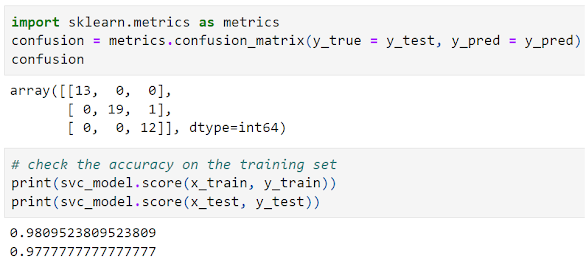
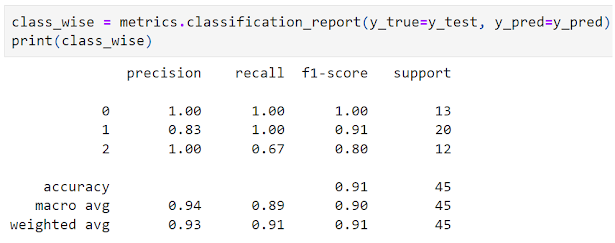
- Model Evaluation
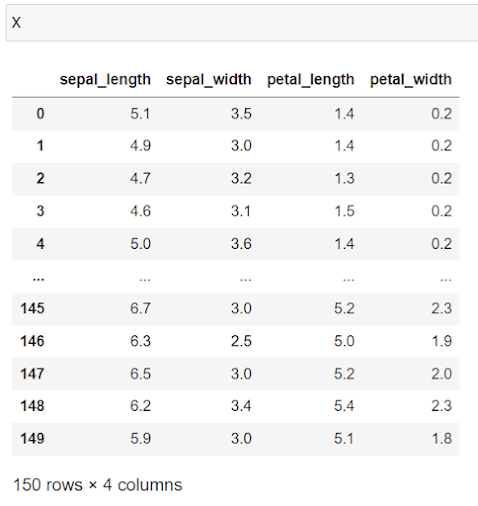
2. Read the dataset (Iris Dataset) and analyze the data
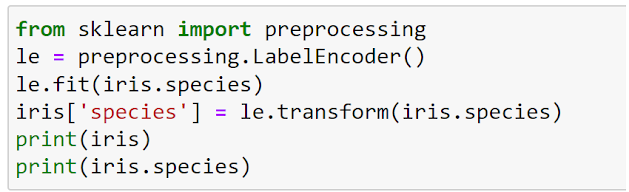
3. Preprocessing the data
4. Divide the data into Training and Testing
5. Build the model - SVM Classifier with different types of kernels
Support vector machines (SVMs) are a set of supervised learning methods used for classification, regression and outliers detection.
The advantages of support vector machines are:
Effective in high dimensional spaces.
Still effective in cases where number of dimensions is greater than the number of samples.
Uses a subset of training points in the decision function (called support vectors), so it is also memory efficient.
Versatile: different Kernel functions can be specified for the decision function. Common kernels are provided, but it is also possible to specify custom kernels.
The disadvantages of support vector machines include:
If the number of features is much greater than the number of samples, avoid over-fitting in choosing Kernel functions and regularization term is crucial.
SVMs do not directly provide probability estimates, these are calculated using an expensive five-fold cross-validation.
class sklearn.svm.SVC(*, C=1.0, kernel='rbf', degree=3, gamma='scale', coef0=0.0, shrinking=True, probability=False, tol=0.001, cache_size=200, class_weight=None, verbose=False, max_iter=-1, decision_function_shape='ovr', break_ties=False, random_state=None)
Regularization parameter. The strength of the regularization is inversely proportional to C. Must be strictly positive. The penalty is a squared l2 penalty.
kernel{‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, ‘precomputed’} or callable, default=’rbf’
Specifies the kernel type to be used in the algorithm. If none is given, ‘rbf’ will be used. If a callable is given it is used to pre-compute the kernel matrix from data matrices; that matrix should be an array of shape
(n_samples, n_samples).degreeint, default=3
Degree of the polynomial kernel function (‘poly’). Ignored by all other kernels.
gamma{‘scale’, ‘auto’} or float, default=’scale’
Kernel coefficient for ‘rbf’, ‘poly’ and ‘sigmoid’.
- if
gamma='scale'(default) is passed then it uses 1 / (n_features * X.var()) as value of gamma,- if ‘auto’, uses 1 / n_features.
- if
gamma changed from ‘auto’ to ‘scale’.fit, will slow down that method as it internally uses 5-fold cross-validation, and predict_proba may be inconsistent with predict.n_samples / (n_classes * np.bincount(y)).decision_function_shape='ovr', and number of classes > 2, predict will break ties according to the confidence values of decision_function; otherwise the first class among the tied classes is returned. Please note that breaking ties comes at a relatively high computational cost compared to a simple predict.probability is False. Pass an int for reproducible output across multiple function calls.